Bacterial Genetics and Genomics book Discussion Topic: Chapter 5, question 13
The regulation of gene expression means that genes are not ‘on’ all of the time, being transcribed and translated to make protein. The orchestration of gene expression is essential to the processes of the bacterial cell and can involve a number of mechanisms.
Often a regulatory protein is involved in the regulation of its own gene. This means that there is a feedback loop where transcription and translation of the gene circles back and regulates its own transcription and translation. Most often we see this happen directly on the promoter of the regulatory gene, where its own protein interacts with the promoter and either represses or activates its own expression. In fact, half of the regulators in E. coli are autoregulated.
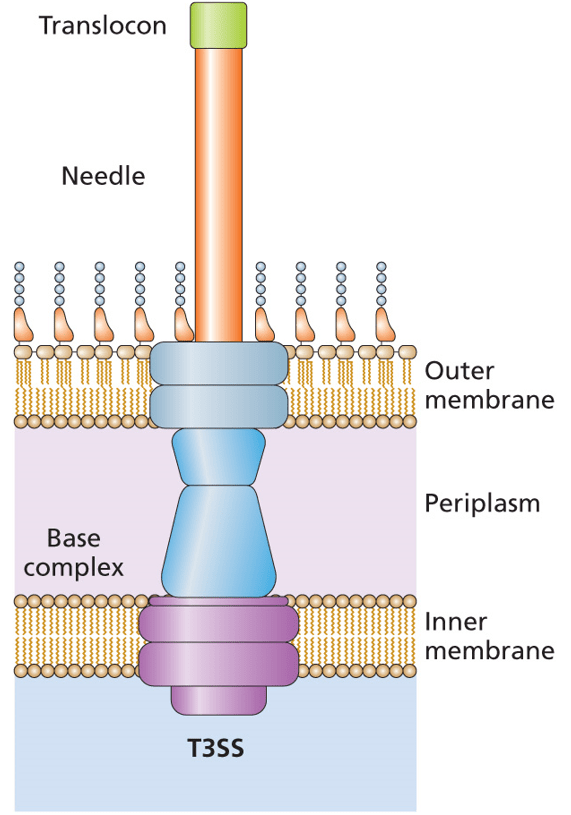
A recent publication describes things a bit differently. In Vibrio parahaemolyticus, the Type 3 Secretion System (T3SS) is regulated by the VtrB activator. These bacteria are resident in the marine environment, where they infect seafood that is part of our food chain. Eating raw or undercooked seafood can lead to an infection with V. parahaemolyticus. The Type 3 Secretion System (actually the second of two T3SSs) is the major virulence factor in these bacteria and it is activated by the VtrB regulatory protein. You can read more about Type 3 Secretion Systems in Chapter 9 and Chapter 13 of Bacterial Genetics and Genomics.
The expression of vtrB is activated by the bile acids that are secreted into the human intestinal tract. This causes the VtrA protein to bind to the promoter region of vtrB, activating its transcription and translation. It has been shown that this activation and expression of VtrB is crucial to infection in humans. Thus understanding it might give insights that could lead to better outcomes for V. parahaemolyticus infections.
The vtrB gene is autoregulated in an interesting way. Rather than VtrB interacting with the promoter of its own gene, the protein is involved in activation of the genes upstream (5’) of vtrB. There is an intrinsic transcriptional terminator between these upstream genes and vtrB. You can read more about intrinsic transcriptional terminators in Chapter 2 of Bacterial Genetics and Genomics.
VtrB causes the transcription of upstream genes to continue through to vtrB, despite the presence of the transcriptional terminator. This read-through transcription increases the expression of VtrB and thus increases the expression of the Type 3 Secretion System in the human body and the associated virulence.

This means that unlike the Type 3 Secretion Systems in species such as Salmonella, Shigella flexneri, and E. coli that autoregulate, with the regulatory protein working with its own promoter, the V. parahaemolyticus relies on read-through transcription. Assessment of the transcriptional terminator suggests that it is weak, with a low stability hairpin being formed, which is less likely to disengage the RNA polymerase. In addition, the transcriptional terminator is also very close (one base away) to the stop codon, which can also lead to read-through. A lesson to take from this is that autoregulation of a transcriptional regulator may be occurring, even if the regulator is not activating or repressing its own promoter. Check upstream of the regulatory protein gene. If the upstream genes are transcribed in the same direction and the transcriptional terminator between is weak, then maybe investigating read-through transcription will be worthwhile.
